Bayesian Segmentation with Histological Atlas
This functionality is available on development versions newer than October 26th 2023
Author: Juan Eugenio Iglesias
E-mail: jiglesiasgonzalez [at] mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
The manuscript describing this module is still in preparation:
Contents
- General Description
- Installation
- Usage
- Frequently asked questions (FAQ)
1. General Description
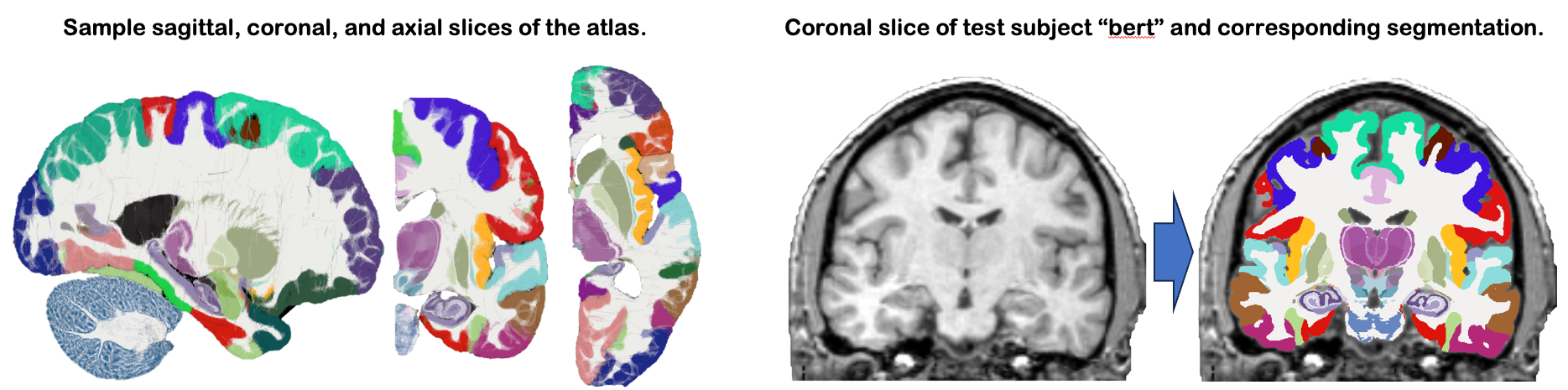
This module uses our new probabilistic atlas of the human brain to segment 333 distinct ROIs per hemisphere on in vivo scans. Segmentation relies on a Bayesian algorithm and is thus robust against changes in MRIi pulse sequence (e.g., T1-weighted, T2-weighted, FLAIR, etc). Sample slices of the atlas and the segmentation of the sample subject "bert" are shown below:
2. Installation
The first time you run this module, it will prompt you to download the atlas. Follow the instructions on the screen to obtain the files.
3. Usage
mri_histo_atlas_segment
4. Frequently asked questions (FAQ)
blah blah
