Bayesian Segmentation with Histological Atlas "NextBrain"
To use this functionality before the next version of FreeSurfer comes out, please install the latest stable version of FreeSurfer (7.4.1), download the code+atlas from this link and follow the instruction in the README file.
Also, please visit the homepage of the NextBrain project for further information.
Author: Juan Eugenio Iglesias
E-mail: jiglesiasgonzalez [at] mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
A preprint of the manuscript describing the atlas and segmentation method used in this module can be found on bioRxiv.
Contents
- General Description
- Installation
- Usage
- Frequently asked questions (FAQ)
1. General Description
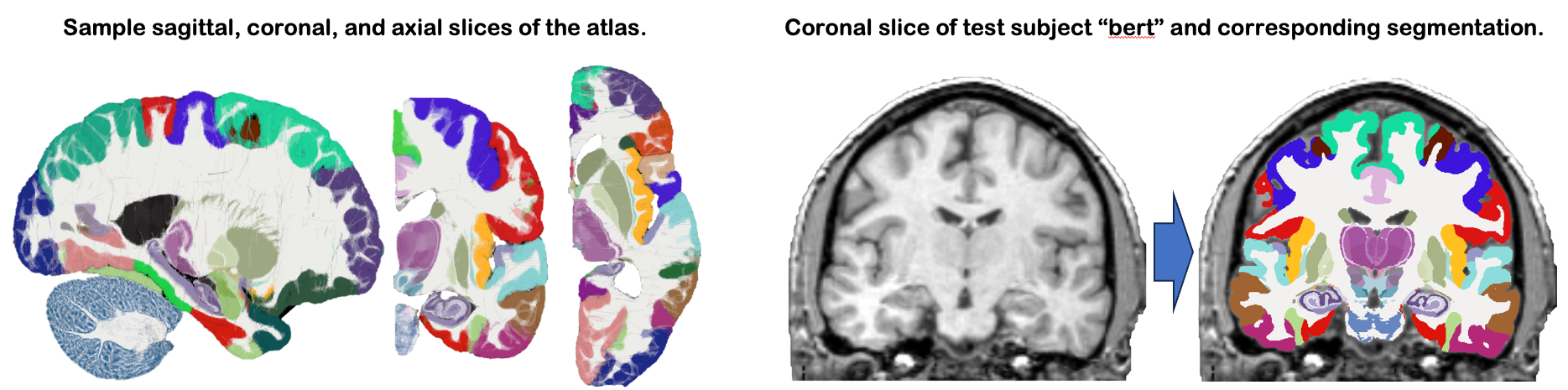
This module uses our new probabilistic atlas of the human brain to segment 333 distinct ROIs per hemisphere on in vivo scans. Segmentation relies on a Bayesian algorithm and is thus robust against changes in MRIi pulse sequence (e.g., T1-weighted, T2-weighted, FLAIR, etc). Sample slices of the atlas and the segmentation of the sample subject "bert" are shown below:
2. Installation
The first time you run this module, it will prompt you to download the atlas. Follow the instructions on the screen to obtain the files.
3. Usage
To segment a brain MRI scan,
mri_histo_atlas_segment INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS
where:
INPUT_SCAN: scan to process, in mgz or nii(.gz) format.
OUTPUT_DIRECTORY: directory where segmentations, volume files, etc, will be created (more on this below).
GPU: set to 1 to use the GPU (we highly recommend using a GPU to run this module; without a GPU, running this module on a single scan can take a whole day). The GPU requirements depend on the image but are about 24GB of memory.
THREADS: number of CPU threads used by the code (set to -1 to use all available threads).
For example:
mri_histo_atlas_segment $SUBJECTS_DIR/bert/mri/orig.mgz $SUBJECTS_DIR/bert/mri/histo_atlas_segmentation/ 1 8
In the output directory, you will find:
bf_corrected.mgz: a bias field corrected version of the input scan.
SynthSeg.mgz: SynthSeg segmentation of the input (which we use in preprocessing and to initialize Gaussian parameters).
MNI_registration.mgz: EasyReg registration to MNI space, use in preprocessing.
seg_[left/right].mgz: segmentation into 333 ROIs of the left and right hemisphere, respectively.
vols_[left/right].csv: CSV spreadsheet with the volumes of the different ROIs, computed from the posteriors (soft segmentations).
lookup_table.txt: FreeSurfer lookup table mapping label indices to brain anatomy. You need it when visualizing the segmentations with Freeview.
4. Frequently asked questions (FAQ)
Do I really need a GPU?
Technically, no. In practice, yes. On a modern GPU, the code runs in an hour or less. On the CPU, it depends on the number of threads, but it can easily take a whole day.
What are the two additional arguments listed on the command line help?
You should not need to touch these, but the 5th argument (BF_MODE) changes the set of basis functions for bias field correction and you could potentially try tinkering with it if the bias field correction fails (i.e., if bf_corrected.mgz has noticeable bias). The 6th argument is GMM_MODE, which allows you to change the grouping of ROIs into tissue classes (advanced mode!).
The GMM model is crucial as it determines how different brain regions are grouped into tissue types for the purpose of image intensity modeling. This is specified though a set of files that should be found under 'data' in the atlas directory:
data/gmm_components_[GMM_MODE].yaml: defines tissue classes and specificies the number of components of the corresponding GMM
data/combined_aseg_labels_[GMM_MODE].yaml defines the labels that belong to each tissue class
data/combined_atlas_labels_[GMM_MODE].yaml defines FreeSurfer ('aseg') labels that are used to initialize the parameters of each class.
We distribute a GMM_MODE named "1mm" that we have used in our experiments, and which is the default mode of the code. If you want to use your own model, you will need to create another triplet of files of your own (use the 1mm version as template).
