|
Size: 6557
Comment:
|
Size: 6558
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 56: | Line 56: |
| To determine the scaling factor, 22 subjects for which the TIV has been determined manually via their T2-weighted scans (which clearly show the skull), are used. These subjects are found in {{{/autofs/space/jc_001/users/nicks/subjects/SASHA}}} (a backup copy is at {{{/autofs/space/tinia_001/users/nicks/subjects/SASHA}}}. The scaling factor is determined analytically by first generating the determinant of the atlas transform for the 22 subjects, plotting this against each subjects manual TIV, fitting a line to this, and calculating the scale factor from the slope. | To determine the scaling factor, 22 subjects for which the TIV has been determined manually via their T2-weighted scans (which clearly show the skull), are used. These subjects are found in {{{/autofs/space/jc_001/users/nicks/subjects/SASHA}}} (a backup copy is at {{{/autofs/space/tinia_001/users/nicks/subjects/SASHA}}}). The scaling factor is determined analytically by first generating the determinant of the atlas transform for the 22 subjects, plotting this against each subjects manual TIV, fitting a line to this, and calculating the scale factor from the slope. |
eTIV - estimated Total Intracranial Volume, aka ICV
Intro
The estimated total intracranial volume (ICV) calculation is based on this work:
A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume Buckner et al. (2004) NeuroImage 23:724-738.
Basically, total intracranial volume is found to correlate with the determinant of the transform matrix used to align an image with an atlas. The work demonstrates that a one-parameter scaling factor provides a reasonable TIV estimation.
Usage
The Freesurfer binaries that calculate eTIV are mri_segstats and mri_label_volume.
recon-all automatically calls mri_segstats, putting the eTIV (also called ICV) in the <subj>/stats/aseg.stats file. Here is an example output line from an aseg.stats file:
# Measure IntraCranialVol, ICV, Intracranial Volume, 2053447.999698, mm^3
mri_segstats hard-codes the transform file used to extract the determinant, and hard-codes the scale factor (see Methods section below for details). To have mri_segstats output just the eTIV:
mri_segstats --subject subjid --etiv-only
In constrast, mri_label_volume requires specifying the transform file to use, and the scale factor:
mri_label_volume -eTIV \ $sdir/transforms/talairach.xfm 1948 \ $sdir/aseg.mgz 17 53
where $sdir is the path to the subject's mri directory (ie, $SUBJECTS_DIR/<subjid>/mri). The '17' and '53' are label ID's, in this case left and right hippocampus (see the subjects /stats/aseg.stats file for IDs). The output will look like:
using eTIV from atlas transform of 1528 cm^3 processing label 17... 3822 voxels (3822.0 mm^3) in label 17, %0.250119 of eTIV volume (1528075) processing label 53... 4410 voxels (4410.0 mm^3) in label 53, %0.288598 of eTIV volume (1528075)
Here, the eTIV is 1528075mm^3.
To check the quality of the registration file talairach.xfm file:
cd $SUBJECTS_DIR/subject/mri/transforms tkregister2 --xfm talairach.xfm \ --targ $FREESURFER_HOME/average/RB_all_withskull_2008-03-26.gca \ --mov ../nu_noneck.mgz --reg junk
Methods
The Buckner et al. paper, referenced above, demonstrates that atlas normalization using appropriate template images provides an automated method for head size correction that is equivalent to manual TIV correction. To implement this work in Freesurfer, three elements are necessary: an appropriate template atlas, normalization to that template, and a scaling factor. In Freesurfer, there are at least two options for a 'template atlas', and accompanying 'normalization' (or transform) to that atlas. One option, used in Freesurfer v4.2.0 and earlier, is to use the atlas file $FREESURFER_HOME/average/RB_all_withskull_2008-03-26.gca and the registration to that atlas found in $SUBJECTS_DIR/subjid/mri/transforms/talairach_with_skull.lta (created by mri_em_register). The other option, that used in later versions of Freesurfer, is to use the atlas $FREESURFER_HOME/average/711-2C_as_mni_average_305.4dfp.img and the registration to that atlas found in $SUBJECTS_DIR/subjid/mri/transforms/talairach.xfm (created by talairach_avi, a script which calls imgreg_4dfp). Note that the '711-2C' target image includes skull (the lack of '_with_skull' in the filename talairach.xfm is a bit misleading).
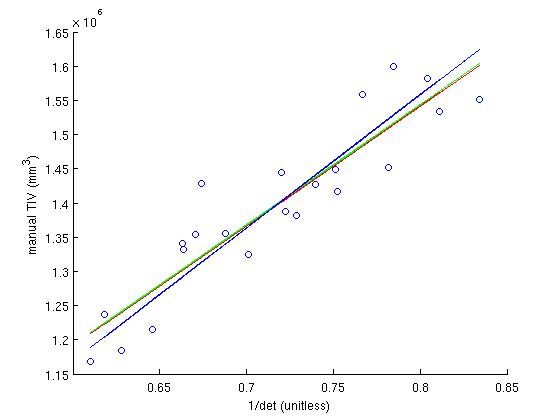
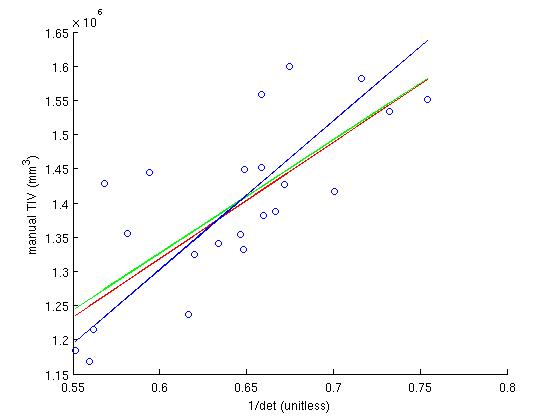
To determine the scaling factor, 22 subjects for which the TIV has been determined manually via their T2-weighted scans (which clearly show the skull), are used. These subjects are found in /autofs/space/jc_001/users/nicks/subjects/SASHA (a backup copy is at /autofs/space/tinia_001/users/nicks/subjects/SASHA). The scaling factor is determined analytically by first generating the determinant of the atlas transform for the 22 subjects, plotting this against each subjects manual TIV, fitting a line to this, and calculating the scale factor from the slope.
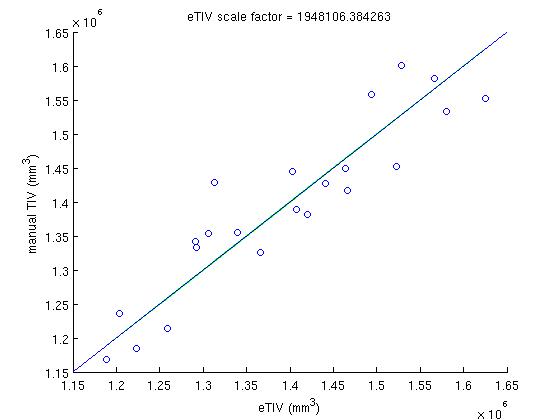
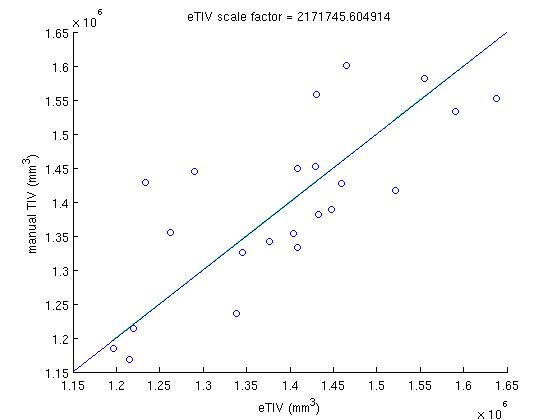
In that SASHA directory, in the the scripts directory, the script run_rb_xfm.csh will run the mri_label_volume binary and generate a matlab data file called det_eTIV_matdat.m which contains the determinant and eTIV data. Then, the matlab script plot_inv_det.m plots the inverse determinant against the manual TIV, finds the best fit to this plot, calculates the scale factor from that fit and then plots the eTIV against manual TIV, for each subject using that scaling factor. The file ICVnative_matdat.m contains the manual TIV data, copied from the ICVnative column in the file buckner_tiv.txt, which originates from the people performing the manual TIV measurements.
new method (post v4.2.0): talairach.xfm |
old method: talairach_with_skull.lta |
|
|
|
|
error: max=8.1%, mean=3.1%, std=1.6%, cv=0.53 |
error: max=13.6%, mean=4.9%, std=3.4%, cv=0.69 |
In the inverse determinant plots, the blue line is regression without y-intercept, and is used to determine the factor. For comparison, the green line is regression and the red line is robust regression with y intercept. The eTIV-manualTIV plots shows the comparison of the estimated and manual TIV's, were we hope to see the identity.
The eTIV was also assessed on two sets of longitudinal scans: a control and a patient undergoing atrophy. The scans were taken on three different platforms (Sonato and Avanto 1.5T and Trio 3T) and two different software versions (vb13 and vb15), over a period of six years. The max absolute relative error (in percent) and the average relative error were found to be:
- control:
- all ten scans (Avanto and Trio): max 4.3%, mean 3.6%
Avanto vb13 & vb15 (seven scans): max 5.1%, mean 2.9%
- Avanto vb15 (five scans): max 1% , mean 0.6%
- patient:
- all 17 scans (Sonato, Avanto and Trio), from 2002 to 2008: max 3%, mean 1%
- Sonata and Avanto vb13 and vb15 (16 scans), from 2002 to 2008: max 2.6%, mean 0.9%
- Avanto vb13 and vb15 (13 scans), from 2004 to 2008: max 0.75%, mean 0.3%
This shows that even in the case of a patient known to be undergoing extensive atrophy, the eTIV variability is small and on par with the control.