|
Size: 2708
Comment:
|
Size: 4591
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from ProjectNameOrWhatever | |
| Line 7: | Line 6: |
<<TableOfContents(5)>> |
|
| Line 14: | Line 15: |
| Usage: ./bundleAnalysis.sh <Patient File> <OPTION: DTI> | Usage: {{{ ./bundleAnalysis.sh <Patient File> <OPTION: DTI> }}} |
| Line 18: | Line 21: |
| This program runs both dmri_groupByEndpoints and dmri_extractSurfaceMeasurements and the outputs will be under its respective section below |
|
| Line 20: | Line 25: |
| === Individually === | == Individually Programs == |
| Line 22: | Line 27: |
| ==== Group by Endpoints ==== | === Group by Endpoints === |
| Line 24: | Line 29: |
| Usage: ./dmri_groupByEndpoints -s <streamlineFile> -i <imageFile> -d <outputDirectory> | Usage: {{{ ./dmri_groupByEndpoints -s <streamlineFile> -i <imageFile> -d <outputDirectory> }}} |
| Line 26: | Line 33: |
| The program will take in a streamline file and an image file, cluster the streamlines, and output TRK files to the output directory | The program will take in a streamline file and an image file, cluster the streamlines based on location, and output TRK files to the output directory |
| Line 28: | Line 35: |
| ===== Output ===== | ==== Output ==== |
| Line 30: | Line 37: |
| ==== Extract Surface Measurements ==== | The output directory will be filled with TRK files. Each TRK file will be labeled with the name of the region and each will be filled with streamlines that begin and end within it. See below: |
| Line 32: | Line 39: |
| Usage: ./dmri_extractSurfaceMeasurements -i <streamlineFile(s)> -sl <Left Hemisphere Surface> -tl <Left Thickness File> -cl <Left Curvature File> -sr <Right Hemisphere Surface> -tr <Right Thickness File> -cl <Right Curvature File> -o <outputDirectory> | {{attachment:Clustered Streamlines.png||width="500",height="350"}} {{attachment:Clustered Streamlines 2.png||width="450",height="350"}} === Extract Surface Measurements === Usage: {{{ ./dmri_extractSurfaceMeasurements -i <streamlineFile(s)> -sl <Left Hemisphere Surface> -tl <Left Thickness File> -cl <Left Curvature File> -sr <Right Hemisphere Surface> -tr <Right Thickness File> -cl <Right Curvature File> -o <outputDirectory> |
| Line 35: | Line 47: |
| }}} | |
| Line 38: | Line 51: |
| ===== Output ===== | ==== Output ==== |
| Line 40: | Line 53: |
| === Additional Programs === | The output directory will be filled with CSV files. Each CSV file will be labeled with the name of the region and each will contain numerous metrics, such as thickness and curvature. If the -fa flag is used, an addition 2 or more column will contain the mean FA and standard deviation. See below: |
| Line 42: | Line 55: |
| ==== Change Endpoints ==== | '''With -FA Flag''' |
| Line 44: | Line 57: |
| Usage: ./dmri_changeEndpoints -i <streamlineFile> -sl <Left Hemisphere Surface> -sr <Right Hemisphere Surface> -ol <Left Overlay Filename> -or <Right Overlay Filename> -itk <Itk Volume File> -fs <FS Volume File> | {{attachment:CSV.png||width="950",height="400"}} '''Without Flag''' {{attachment:CSV_noFA.png||width="500",height="400"}} == Additional Programs == === Change Endpoints === Usage: {{{ ./dmri_changeEndpoints -i <streamlineFile> -sl <Left Hemisphere Surface> -sr <Right Hemisphere Surface> -ol <Left Overlay Filename> -or <Right Overlay Filename> -itk <Itk Volume File> -fs <FS Volume File> }}} |
| Line 48: | Line 73: |
| ===== Output ===== | ==== Output ==== |
| Line 50: | Line 75: |
| ==== Plots ==== | The new overlay files will be created in the same directory with the name that it was passed with. The overlay files will be completely blank (values are zero) besides the points near the beginning and end of the streamline passed in, which will have a value that was set before it was run. See below: |
| Line 52: | Line 77: |
| Usage: python3 ./dmri_plots.py <inputDirectory> <Grouping CSV> <Structure Name> | {{attachment:Endpoints.png||width="650",height="350"}} {{attachment:Endpoints 2.png||width="650",height="350"}} |
| Line 54: | Line 79: |
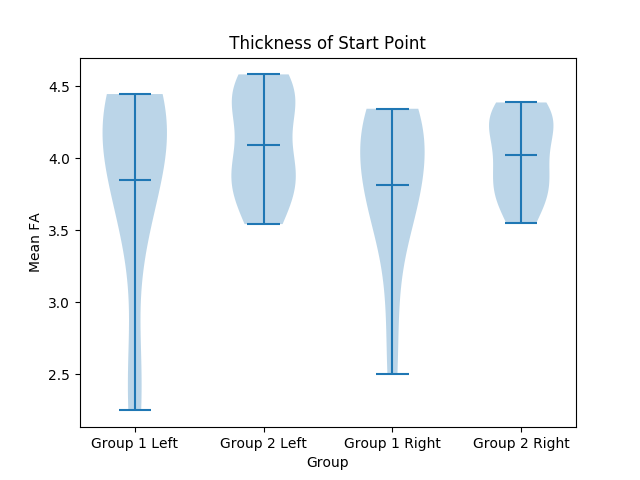
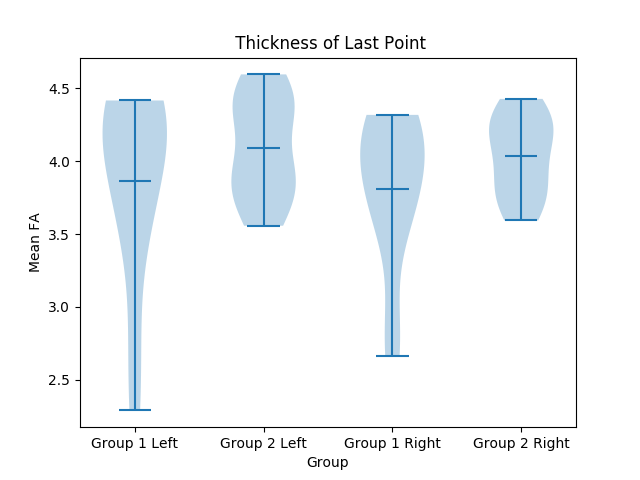
| The program will take in an input Directory (containing subject directories), a CSV files that divides the subjects into groups, and the name of a structure. For every subject, multiple violin plots are created to organize the data from every column of the structure's CSV file. The plot will be further explained below. | === Plots === |
| Line 56: | Line 81: |
| ===== Output ===== | Usage: {{{ python3 ./dmri_plots.py <inputDirectory> <Grouping CSV> <Structure Name> }}} The program will take in an input directory containing the subjects as subdirectories, a CSV file that divides the subjects into groups, and the name of a structure. For every subject, multiple violin plots are created to organize the data from every column of the structure's CSV file. The plot will be further explained below. ==== Output ==== The output will be a number of violin plots based on the amount of value types. In the data, each group is represented by 2 violin plots, one for both the left and right halves. For example, on the left half, these two violins represent the left and right hemisphere of group 1, while the other two represent the left and right hemisphere of group 2. See below: {{attachment:Violin Plot 1.png||width="650",height="300"}} {{attachment:Violin Plot 2.png||width="650",height="300"}} |
U-Shape Bundle Analysis
Note: all of the source code can be found on the "dmri" branch on the Freesurfer Github page. The link can be found here:
https://github.com/freesurfer/freesurfer/tree/dmri/anatomicuts
Contents
How to Run
Note by running the executable with no parameters will also give information on how to run the program
Main Bundle Analysis Program
Usage:
./bundleAnalysis.sh <Patient File> <OPTION: DTI>
DTI is an option that allows more metrics about the FA values to be outputted based on 4 volumes
This program runs both dmri_groupByEndpoints and dmri_extractSurfaceMeasurements and the outputs will be under its respective section below
If specific files are wanted to be used or files cannot be found, run the programs individually (read below)
Individually Programs
Group by Endpoints
Usage:
./dmri_groupByEndpoints -s <streamlineFile> -i <imageFile> -d <outputDirectory>
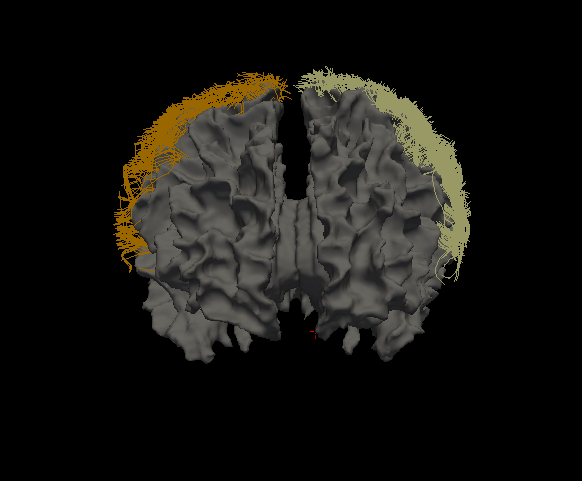
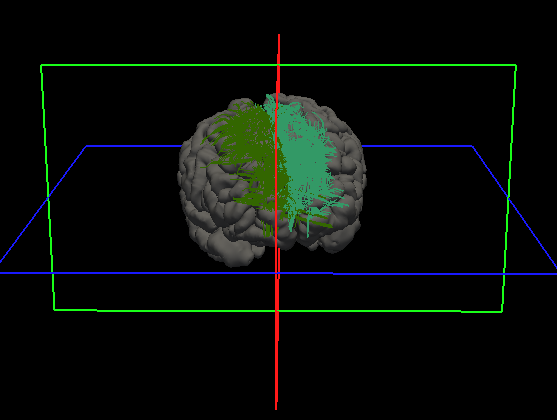
The program will take in a streamline file and an image file, cluster the streamlines based on location, and output TRK files to the output directory
Output
The output directory will be filled with TRK files. Each TRK file will be labeled with the name of the region and each will be filled with streamlines that begin and end within it. See below:
Extract Surface Measurements
Usage:
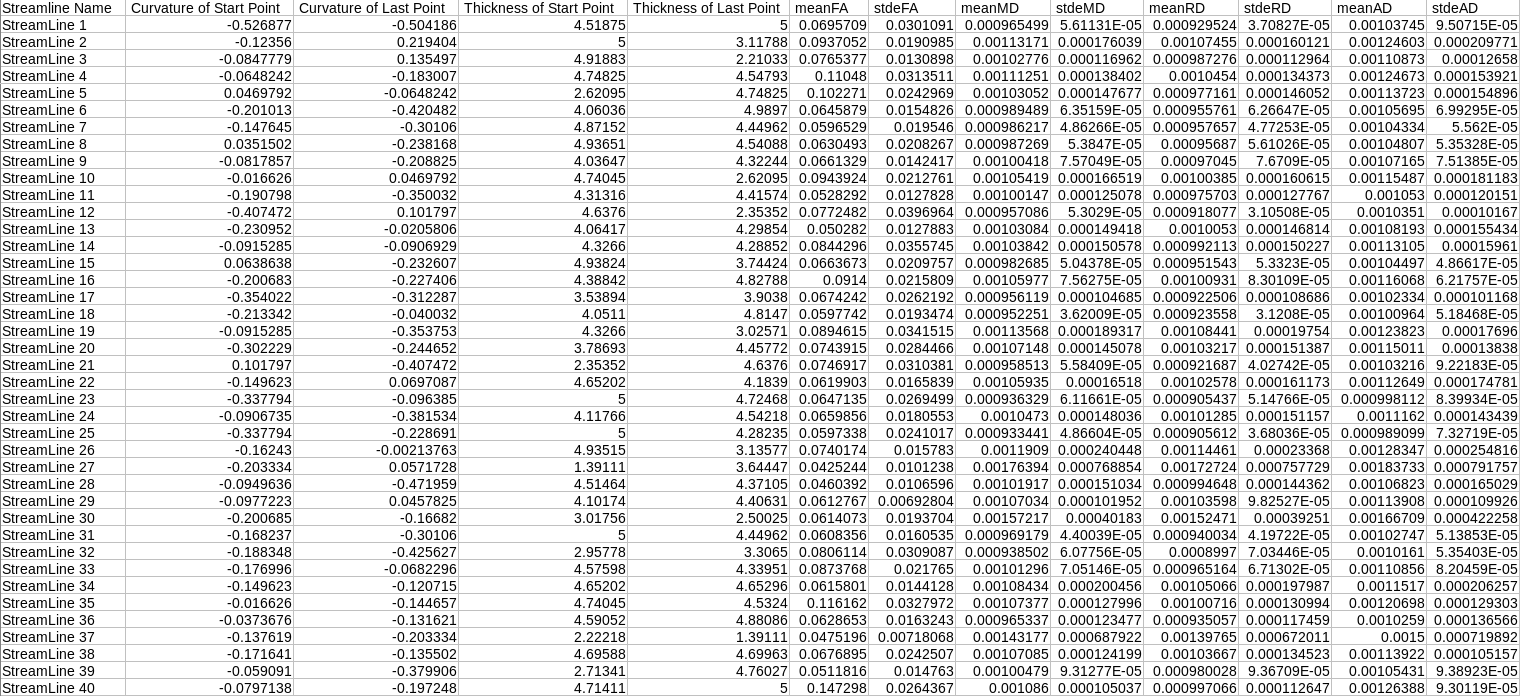
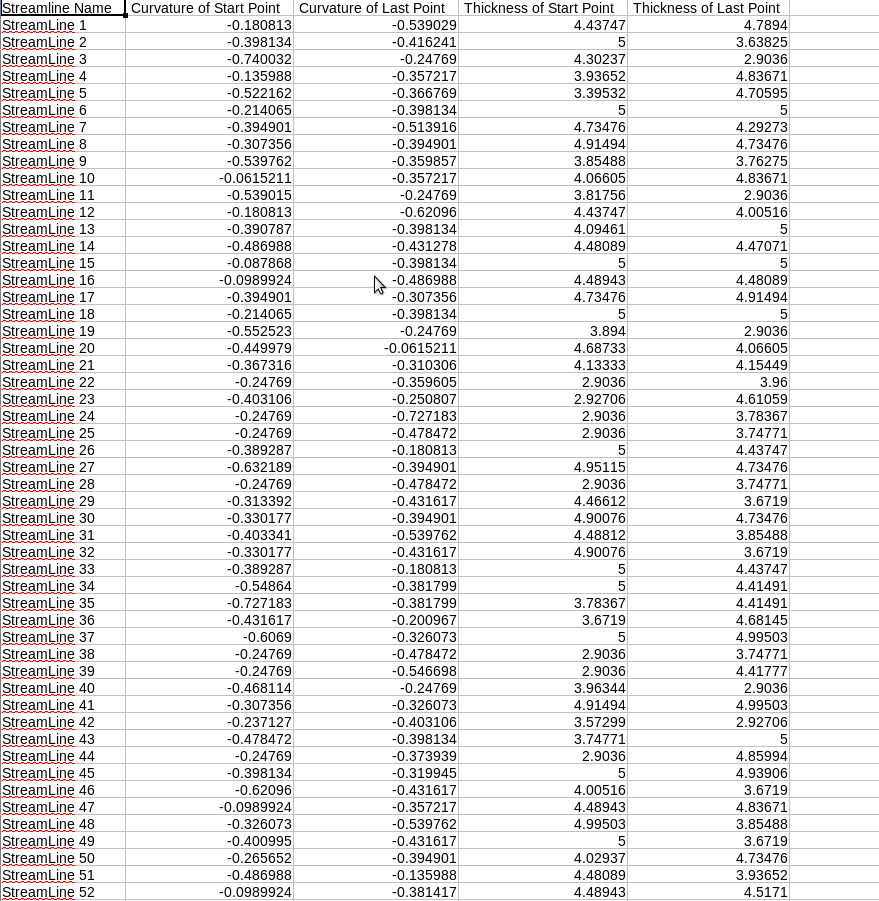
./dmri_extractSurfaceMeasurements -i <streamlineFile(s)> -sl <Left Hemisphere Surface> -tl <Left Thickness File> -cl <Left Curvature File> -sr <Right Hemisphere Surface> -tr <Right Thickness File> -cl <Right Curvature File> -o <outputDirectory> OPTIONAL FLAG: -fa <number of Measures> <Given Name 1> <File 1> ... <Given Name X> <File X>
The program will take in streamlines, a full brain surface with overlay files, an output directory, and possibly image files, and will output surface metrics, such as thickness and curvature as well as FA at the endpoints of the streamlines
Output
The output directory will be filled with CSV files. Each CSV file will be labeled with the name of the region and each will contain numerous metrics, such as thickness and curvature. If the -fa flag is used, an addition 2 or more column will contain the mean FA and standard deviation. See below:
With -FA Flag
Without Flag
Additional Programs
Change Endpoints
Usage:
./dmri_changeEndpoints -i <streamlineFile> -sl <Left Hemisphere Surface> -sr <Right Hemisphere Surface> -ol <Left Overlay Filename> -or <Right Overlay Filename> -itk <Itk Volume File> -fs <FS Volume File>
The program will take in two volumes, a streamline, a surface, and the names of the overlay file that will be created. It will assign the endpoints of the streamline to a set number (usually a value of 1, but can be changed by adjusting the value of the variable at the top)
Output
The new overlay files will be created in the same directory with the name that it was passed with. The overlay files will be completely blank (values are zero) besides the points near the beginning and end of the streamline passed in, which will have a value that was set before it was run. See below:
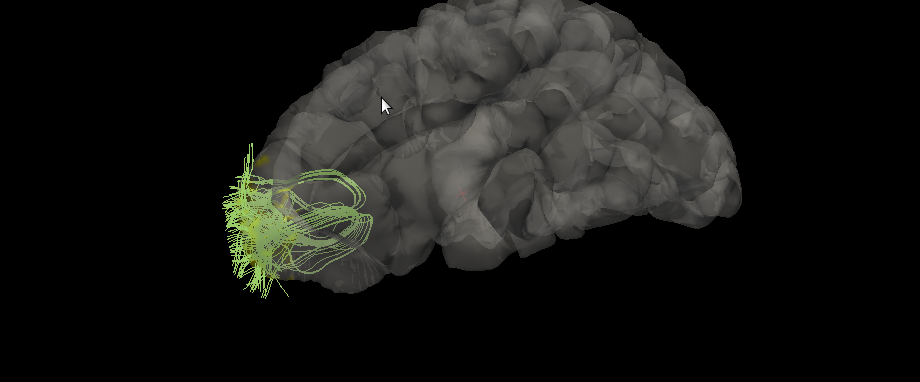
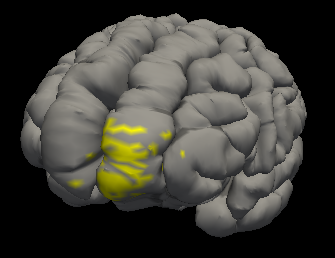
Plots
Usage:
python3 ./dmri_plots.py <inputDirectory> <Grouping CSV> <Structure Name>
The program will take in an input directory containing the subjects as subdirectories, a CSV file that divides the subjects into groups, and the name of a structure. For every subject, multiple violin plots are created to organize the data from every column of the structure's CSV file. The plot will be further explained below.
Output
The output will be a number of violin plots based on the amount of value types. In the data, each group is represented by 2 violin plots, one for both the left and right halves. For example, on the left half, these two violins represent the left and right hemisphere of group 1, while the other two represent the left and right hemisphere of group 2. See below: