| Deletions are marked like this. | Additions are marked like this. |
| Line 53: | Line 53: |
| {{attachment:QA.png||width="500"}} | {{attachment:QA.png||height="350"}} |
Introduction
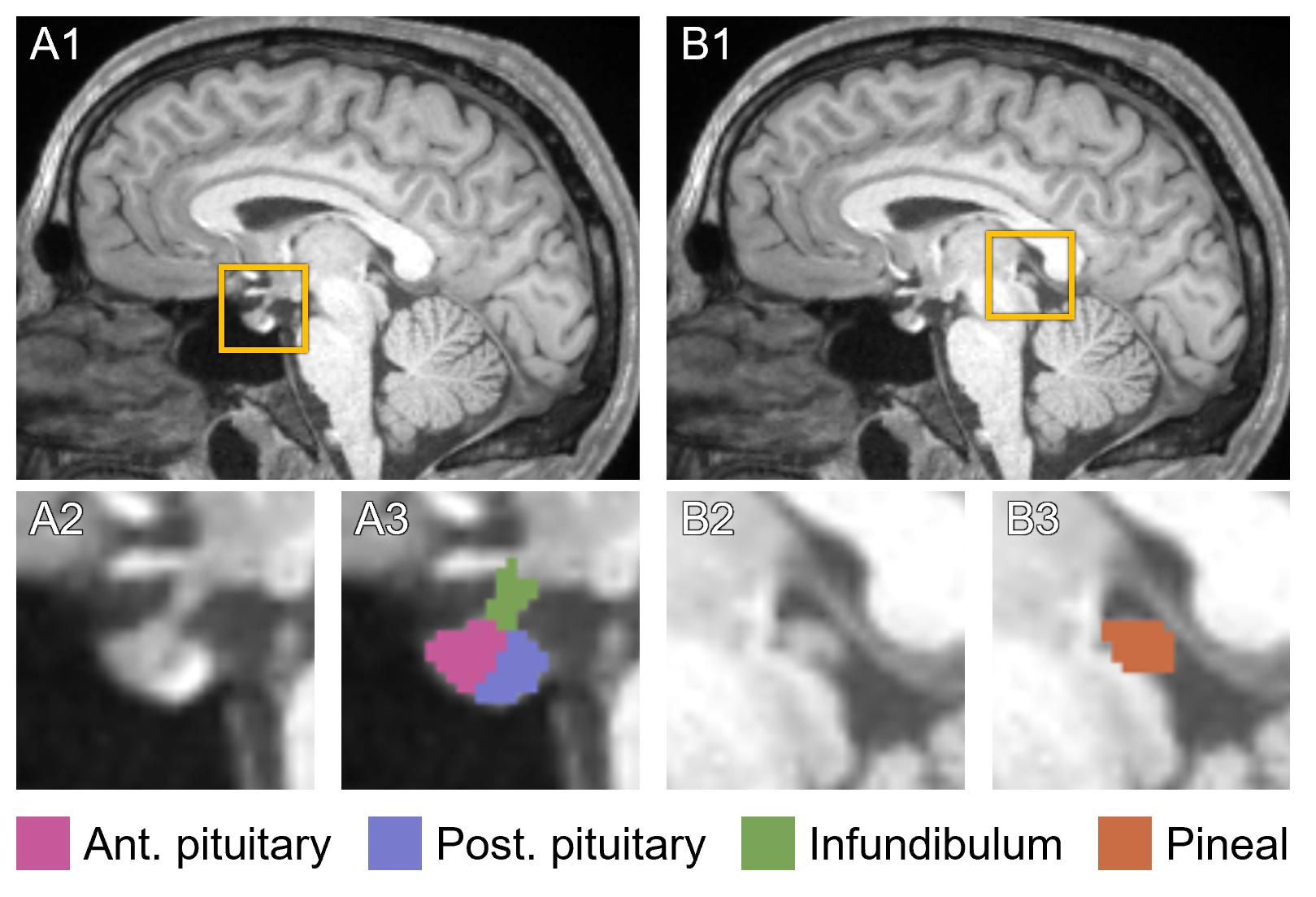
This page describes mri_pglands_seg, a deep learning tool within FreeSurfer to automatically segment the pituitary and pineal glands in a T1-weighted image. Specifically, the pipeline target four structures: the anterior pituitary, posterior pituitary, infundibulum, and pineal gland (see below).
The package requires installation of FreeSurfer version 8.1 or later.
Those using this toolbox should cite: Automated Segmentation of the Pituitary and Pineal Glands. Larson, KL, Augustinack, JC, Mora, J, Shahzade, D, Rapalino, O, Fischl, B, Greve, DN. Human Brain Mapping 46, no. 15: e70384. 10.1002/hbm.70384. https://doi.org/10.1002/hbm.70384. PDF available here.
Basic Usage
There are two ways to run mri_pglands_seg, referred to as "normal mode" and "FreeSurfer mode". In normal mode, the user can specify one or more T1-weighted input images and either a single output directory or a corresponding list of output paths:
mri_pglands_seg --i <input1> ... <inputN> --o <output_dir>
or
mri_pglands_seg --i <input1> ... <inputN> --o <output1> ... <outputN>
If specifying a single input directory instead of a list of images, mri_pglands_seg will attempt to perform segmentation on all files within that directory with valid file extensions (e.g., .mgz, .mgh, .nii, .nii.gz).
Alternatively, FreeSurfer mode allows the user to perform segmentation on subjects processed with the recon-all pipeline, and assumes the standard filename conventions. Instead of supplying a list of inputs and outputs, the user can specify a list of subjects and the SUBJECTS_DIR environment variable (if not already set).
mri_pglands_seg --s <subjectid1> ... <subjectidN> --sd <SUBJECTS_DIR>
The output label maps for each subject will automatically be written to <SUBJECTS_DIR>/<subjectid>/mri/T1.pglands.mgz. The user can also supply an output directory as in normal mode; this will fource all outputs to be written to <output_dir>/<subjectid>.T1.pglands.mgz.
Volume stats
To record the volumes (mm3) of each segmented structure, run mri_pglands_seg with the --write_vol_stats flag, e.g.:
mri_pglands_seg --s <subjectid> --write_vol_stats
This will produce a table in the standard .stats file format with the label value, estimated volume, and number of voxels for each label. The user can also specify --etiv to include the estimated total intracranial volume for that subject within the output stats file. This is automatically included when running in FS mode.
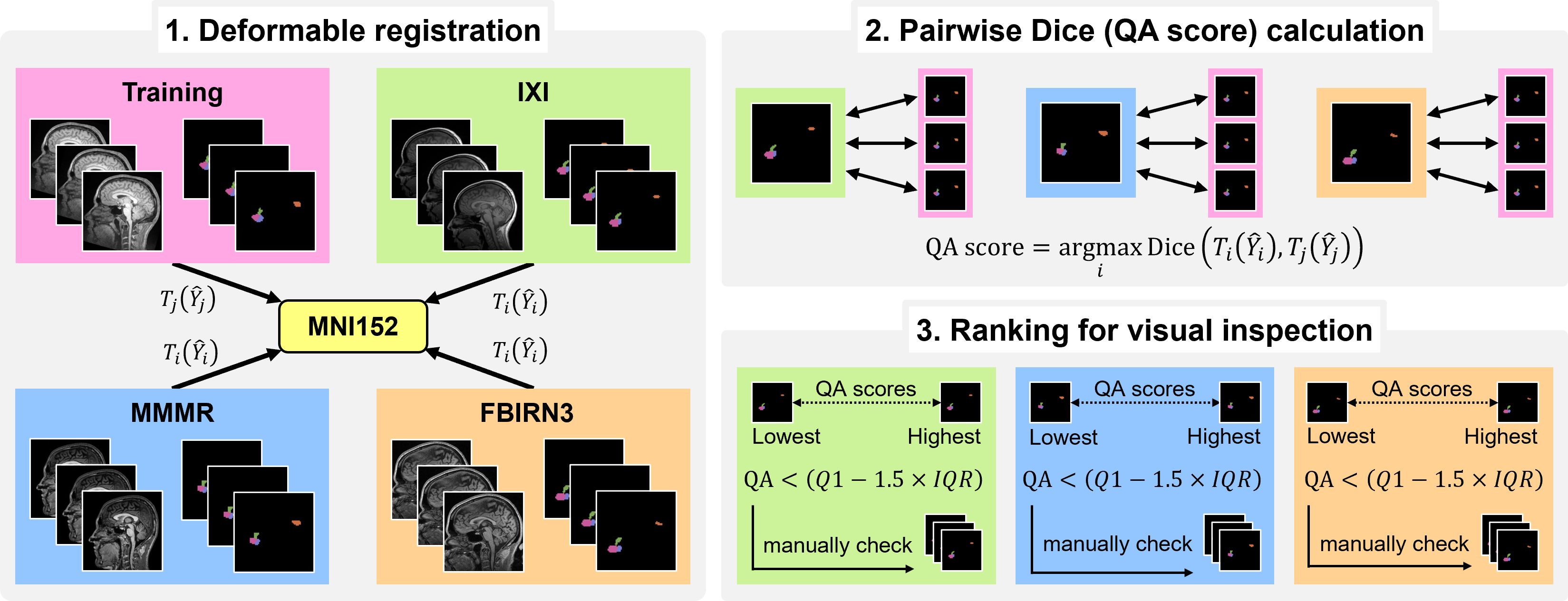
Quality Assurance
The best way to assess the quality of predicted segmentations is to visualize all outputs using the freeview command within FreeSurfer (or your favorite viewing program). Because it can be tedious to view every single result for large datasets, we incorporated a quality assurance (QA) procedure that automatically assigns a "QA score" indicating the accuracy of each segmentations. To run this, use the --write_qa_stats flag:
mri_pglands_seg --s <subjectid> --write_qa_stats
Below is a diagram overviewing our QA procedure. A detailed description is contained within the associated journal article.