|
Size: 3609
Comment:
|
Size: 4997
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [[FsTutorial/MultiModal|Back to Multimodal Top]] cd $TUTORIAL_DATA/multimodal/ |
[[FsTutorial|Back to Top Tutorial Page]]<<BR>> [[FsTutorial/MultiModal|Back to Multimodal Top]] Other multimodal tutorials: <<BR>> [[FsTutorial/MultiModalRegistration|A. Multimodal Registration]], [[FsTutorial/MultiModalFmriIndividual|B. Individual fMRI Integration]], [[FsTutorial/MultiModalFmriGroup|C. Surface-based Group fMRI Analysis]]<<BR>> |
| Line 4: | Line 3: |
[[FsTutorial/MultiModal|Back to Multimodal Top]] The purpose of this tutorial is to give you experience with the integration of Diffusion Tensor Imaging (DTI) with FreeSurfer. The data were collected at MGH as part of the MIND. |
The purpose of this tutorial is to give you experience with the integration of Diffusion Tensor Imaging (DTI) with FreeSurfer. The data were collected at MGH as part of the MIND. |
| Line 12: | Line 6: |
| It is assumed that you already have knowledge of DTI and its analysis. This paragraph is supplied as a summary for completeness. DTI attempts to measure the diffusion of water in the brain. White matter tracts tend to be like little straws which constrain the direction diffusion. The DTI analysis measures the orientation and the "strength" of the orientation. Eg, CSF will have no strong orientation. This strength is often measured as the fractional anisotropy (FA). The diffusivity is often measured as the Apparent Diffusion Coefficient (ADC). | |
| Line 13: | Line 8: |
| It is assumed that you already have knowledge of DTI and its analysis. This paragraph is supplied as a summary for completeness. DTI attempts to measure the diffusion of water in the brain. White matter tracts tend to be like little straws which contrain the direction diffusion. The DTI analysis measures the orientation and the "strength" of the orientation. Eg, CSF will have no strong orientation. This strength is often measured as the fractional anisotropy (FA). The diffusivity is ofen measured as the Apparent Diffusion Coefficient (ADC). |
Prior to DTI analysis, it is customary to motion correct, and sometimes eddy current correct, the diffusion weighted images. This is done by selecting one of the images as a template (usually a low-b volume). This forces all the DTI-related maps to be in-line with the template. Consequently, this template should be used as the registration target. |
| Line 24: | Line 11: |
| This data was acquired at MGH as part of a MIND Consortium study. The raw data are in dwi.nii.gz. The FreeSurfer anatomical analysis for this subject is M87102113.v4. Seventy images were acquired (10 low-b and 60 diffusion weighted). The bvalues and gradient directions are stored in bvals.dat and bvects.dat, respectively. These data were analyzed with FreeSurfer's [[dt_recon]] program, which created the fa.nii, adc.nii, lowb.nii, and register.dat. The data are located in: | |
| Line 25: | Line 13: |
| This data was acquried at MGH as part of a MIND Consortium study. The raw data are in dwi.nii.gz. The FreeSurfer anatomical analysis for this subject is M87102113.v4. Seventy images were acquired (10 low-b and 60 diffusion weighted). The bvalues and gradient directions are stored in bvals.dat and bvects.dat, respectively. These data were analyzed with FreeSurfer's [[dt_recon|dt_recon]] program, which created the fa.nii, adc.nii, lowb.nii, and register.dat. The data are located in: |
---- |
| Line 35: | Line 15: |
| setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs | |
| Line 37: | Line 18: |
| ---- | |
| Line 39: | Line 21: |
| As always, check the registration with the template. In this case, the template is the low-b image: | |
| Line 40: | Line 23: |
| As always, check the registration with the template. In this case, the template is the low-b image: |
---- |
| Line 46: | Line 27: |
The registration should be accurate. For more information on registration, see the [[MultiModalRegistration|Registration]] Tutorial. |
---- The registration should be accurate. For more information on registration, see the [[FsTutorial/MultiModalRegistration|Registration]] Tutorial. |
| Line 52: | Line 31: |
| View the FA on the subject's anatomical with the white matter parcellation (wmparc.mgz) as the segmentation: | |
| Line 53: | Line 33: |
| View the FA on the subject's anatomical with the white matter parcellation (wmparc.mgz) as the segmentation: |
---- |
| Line 57: | Line 35: |
| tkmedit M87102113.v4 orig.mgz -aux brain.mgz \ | tkmedit xxxxxxxxxxx.v4 orig.mgz -aux brain.mgz \ |
| Line 60: | Line 38: |
| -fthresh 0.1 -fmax 1 | -fthresh 0.2 -fmax 1 |
| Line 62: | Line 40: |
| ---- Notes: |
|
| Line 63: | Line 43: |
| Notes: 1. The FA is a values between 0 and 1, so the thresholds are set to 0.1 and 1. 1. When the window first comes up, there will be a lot of activity outside the brain. To remove this, set the View->Mask Functional Overlay To AuxVolume flag. 1. In the image below, you can see that white matter tends to have a higher FA. |
1. The FA is a values between 0 and 1, so the thresholds are set to 0.2 and 1. 1. When the window first comes up, there will be a lot of activity outside the brain. To remove this, set the 'View->Mask Functional Overlay To Aux Volume' flag. 1. In the image below, you can see that white matter tends to have a higher FA. |
| Line 70: | Line 48: |
| PIC | {{attachment:fa.cor128.jpg|fa.cor128.gif}} |
| Line 73: | Line 51: |
| As with the [[FsTutorial/MultiModalFmriIndividual|Individual fMRI Analysis]], we will first resample the FA into the subject's anatomical space: | |
| Line 74: | Line 53: |
| As with the [[MultiModalFmriIndividual| Individual fMRI Analysis]], we will first resample the FA into the subject's anatomical space: |
---- |
| Line 83: | Line 60: |
| ---- Notes: |
|
| Line 84: | Line 63: |
| Notes: 1. Nearest neighbor is used to avoid averaging FA across voxels. 2. The output fa.anat.mgh will be 256^3, 1mm3. |
1. Nearest neighbor is used to avoid averaging FA across voxels. 1. The output fa.anat.mgh will be 256^3, 1mm3. |
| Line 88: | Line 66: |
| Now run mri_segstats getting a report for each of the following ROIs (index/name pairs are found in the LUT): |
Now run mri_segstats getting a report for each of the following ROIs (index/name pairs are found in the LUT): |
| Line 92: | Line 70: |
| 3024 wm-lh-precentral | 3024 wm-lh-precentral |
| Line 94: | Line 72: |
| 3021 wm-lh-pericalcarine 4 Left-Lateral-Ventricle |
3021 wm-lh-pericalcarine 12 Left-Putamen 4 Left-Lateral-Ventricle |
| Line 97: | Line 76: |
The wm-lh-precentral, wm-lh-superiortemporal, wm-lh-pericalcarine are created by the gyral white matter parcellation. |
The wm-lh-precentral, wm-lh-superiortemporal, wm-lh-pericalcarine are created by the gyral white matter parcellation. |
| Line 103: | Line 80: |
| ---- | |
| Line 105: | Line 83: |
| --seg $SUBJECTS_DIR/M87102113.v4/mri/wmparc.mgz \ | --seg $SUBJECTS_DIR/xxxxxxxxx.v4/mri/wmparc.mgz \ |
| Line 107: | Line 85: |
| --id 251 --id 3021 --id 3024 --id 3030 --id 4\ | --id 251 --id 3021 --id 3024 --id 3030 --id 12 --id 4 \ |
| Line 110: | Line 88: |
| ---- Click [[FsTutorial/MultiModalFaStats|HERE]] to see the output. The CC has an average FA of about 0.75, gyral parcellations are about 0.4, the left putamen is 0.27, and the ventricle is 0.2. This is as expected because the CC is highly directional with no crossing fibers so we would expect the CC to have the highest FA. The gyral white matter is also directional but has fibers crossing in them, so one expects the FA to be lower than CC. The gray matter (putamen) is still lower. The ventricle has no fibers, so we expect it to have the lowest FA. |
|
| Line 111: | Line 91: |
| Click HERE to see the output. The CC has an average FA of about 0.75, gyral parcellations are about 0.4, and the ventricle is 0.2. |
Other multimodal tutorials: [[FsTutorial/MultiModalRegistration|A. Multimodal Registration]], [[FsTutorial/MultiModalFmriIndividual|B. Individual fMRI Integration]], [[FsTutorial/MultiModalFmriGroup|C. Surface-based Group fMRI Analysis]]<<BR>> |
Back to Top Tutorial Page
Back to Multimodal Top Other multimodal tutorials:
A. Multimodal Registration, B. Individual fMRI Integration, C. Surface-based Group fMRI Analysis
The purpose of this tutorial is to give you experience with the integration of Diffusion Tensor Imaging (DTI) with FreeSurfer. The data were collected at MGH as part of the MIND.
DTI Basics
It is assumed that you already have knowledge of DTI and its analysis. This paragraph is supplied as a summary for completeness. DTI attempts to measure the diffusion of water in the brain. White matter tracts tend to be like little straws which constrain the direction diffusion. The DTI analysis measures the orientation and the "strength" of the orientation. Eg, CSF will have no strong orientation. This strength is often measured as the fractional anisotropy (FA). The diffusivity is often measured as the Apparent Diffusion Coefficient (ADC).
Prior to DTI analysis, it is customary to motion correct, and sometimes eddy current correct, the diffusion weighted images. This is done by selecting one of the images as a template (usually a low-b volume). This forces all the DTI-related maps to be in-line with the template. Consequently, this template should be used as the registration target.
This Data Set
This data was acquired at MGH as part of a MIND Consortium study. The raw data are in dwi.nii.gz. The FreeSurfer anatomical analysis for this subject is M87102113.v4. Seventy images were acquired (10 low-b and 60 diffusion weighted). The bvalues and gradient directions are stored in bvals.dat and bvects.dat, respectively. These data were analyzed with FreeSurfer's dt_recon program, which created the fa.nii, adc.nii, lowb.nii, and register.dat. The data are located in:
setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $TUTORIAL_DATA/multimodal/dti
Check the Registration
As always, check the registration with the template. In this case, the template is the low-b image:
tkregister2 --mov lowb.nii --reg register.dat --surf
The registration should be accurate. For more information on registration, see the Registration Tutorial.
FA Viewing
View the FA on the subject's anatomical with the white matter parcellation (wmparc.mgz) as the segmentation:
tkmedit xxxxxxxxxxx.v4 orig.mgz -aux brain.mgz \ -seg wmparc.mgz \ -reg register.dat -overlay fa.nii \ -fthresh 0.2 -fmax 1
Notes:
- The FA is a values between 0 and 1, so the thresholds are set to 0.2 and 1.
- When the window first comes up, there will be a lot of activity
outside the brain. To remove this, set the 'View->Mask Functional Overlay To Aux Volume' flag.
- In the image below, you can see that white matter tends to have a higher FA.
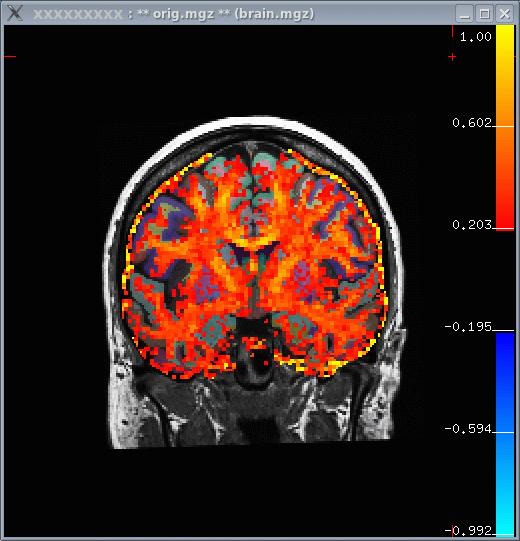
FA ROI Analysis
As with the Individual fMRI Analysis, we will first resample the FA into the subject's anatomical space:
mri_vol2vol --mov fa.nii \ --reg register.dat \ --fstarg --interp nearest \ --o fa.anat.mgh
Notes:
- Nearest neighbor is used to avoid averaging FA across voxels.
- The output fa.anat.mgh will be 256^3, 1mm3.
Now run mri_segstats getting a report for each of the following ROIs (index/name pairs are found in the LUT):
251 CC_Posterior (Posterior Corpus Callosum) 3024 wm-lh-precentral 3030 wm-lh-superiortemporal 3021 wm-lh-pericalcarine 12 Left-Putamen 4 Left-Lateral-Ventricle
The wm-lh-precentral, wm-lh-superiortemporal, wm-lh-pericalcarine are created by the gyral white matter parcellation.
Now run mri_segstats:
mri_segstats \ --seg $SUBJECTS_DIR/xxxxxxxxx.v4/mri/wmparc.mgz \ --ctab $FREESURFER_HOME/FreeSurferColorLUT.txt \ --id 251 --id 3021 --id 3024 --id 3030 --id 12 --id 4 \ --i fa.anat.mgh --sum fa.stats
Click HERE to see the output. The CC has an average FA of about 0.75, gyral parcellations are about 0.4, the left putamen is 0.27, and the ventricle is 0.2. This is as expected because the CC is highly directional with no crossing fibers so we would expect the CC to have the highest FA. The gyral white matter is also directional but has fibers crossing in them, so one expects the FA to be lower than CC. The gray matter (putamen) is still lower. The ventricle has no fibers, so we expect it to have the lowest FA.
Other multimodal tutorials: A. Multimodal Registration, B. Individual fMRI Integration, C. Surface-based Group fMRI Analysis
