|
Size: 1896
Comment:
|
← Revision 14 as of 2011-12-07 13:01:49 ⇥
Size: 2082
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| *To follow this exercise exactly be sure you've downloaded the [[FsTutorial/Data|tutorial data set]] before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. | * To follow this exercise exactly be sure you've downloaded the [[FsTutorial/Data|tutorial data set]] before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. |
| Line 6: | Line 6: |
If preferred, one can use the edited cross sectional data to create a new base. However, edits made to the brain.finalsurfs volume of the cross sectional data sets will not transfer to the base since the base creation takes off from the norm.mgz of the cross data sets. |
Edits made to the brain.finalsurfs volume of the cross sectional data sets will not transfer to the base since the base creation takes off from the norm.mgz of the cross data sets. |
| Line 11: | Line 10: |
| Below is a visualization of the problem area in the base: brain.finalsurfs.mgz, slice 89 at coordinates 98, 172, 89 in right hemi. The problem area extends through slices 85 - 94. <<BR>> {{attachment:basebefore.jpg}} |
Below is a visualization of the problem area in the base: brain.finalsurfs.mgz, slice 89 at coordinates 109 166 89 in right hemi. The problem area extends through slices 85 - 91. <<BR>> {{attachment:baseBEFORE.jpg}} |
| Line 14: | Line 12: |
| And here is the area zoomed in: <<BR>> {{attachment:basebeforezoomed.jpg}} |
And here is the area zoomed in: <<BR>> {{attachment:baseBEFOREzoomed.jpg}} |
| Line 17: | Line 14: |
| Next, edit the affected slices by removing the appropriate voxels, and save the changes to the aux volume. Then make a copy of the edited brain.finalsurfs.mgz file to one called brain.finalsurfs.manedit.mgz in the base: | First make a copy of the brain.finalsurfs.mgz volume to one called 'brain.finalsurfs.manedit.mgz' and save it within the mri directory of the base: |
| Line 19: | Line 17: |
| cp OAS2_0002/mri/brain.finalsurfs.mgz OAS2_0002/mri/brain.finalsurfs.manedit.mgz | cp OAS2_0002/mri/brain.finalsurfs.mgz OAS2_0002/mri/brain.finalsurfs.manedit.mgz |
| Line 21: | Line 19: |
| Next, load the brain.finalsurfs.manedit.mgz volume as the aux volume in tkmedit and edit this volume by going through the affected slices and removing the voxels in the cerebellum and surrounding areas that cause the pial surface misplacement. Once you are finished editing, save the changes to the volume with File – Save Aux volume. Then, run the following to have edits take effect: | |
| Line 22: | Line 21: |
| Next, run the following to have edits take effect: | |
| Line 24: | Line 22: |
| recon-all -base OAS2_0002 -autorecon3-pial | recon-all -base OAS2_0002 -autorecon-pial |
| Line 26: | Line 24: |
| Results will look as follows (see OAS2_0002_fixed): <<BR>> {{attachment:baseFIXED.jpg}} | |
| Line 27: | Line 26: |
| Results will look as follows: <<BR>> {{attachment:basefixed.jpg}} |
Here is zoomed in version of above: <<BR>> {{attachment:baseFIXEDzoomed.jpg}} |
| Line 30: | Line 28: |
| Here is zoomed in version of above: <<BR>> {{attachment:basefixedzoom.jpg}} Now that the base and cross sectionals are fixed, you must re-create the longitudinal data sets for each time point. Return to the [[FsTutorial/LongitudinalTutorial|Longitudina Tutorial]] to read that section. |
Now that the base and cross sectional time point 2 are fixed, you must re-create the longitudinal data set for time point 2. Return to the [[FsTutorial/LongitudinalTutorial|Longitudinal Tutorial]] to read that section. |
Making FinalSurf edits - base
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
Edits made to the brain.finalsurfs volume of the cross sectional data sets will not transfer to the base since the base creation takes off from the norm.mgz of the cross data sets.
Therefore, in order to fix the base, one must edit the brain.finalsurfs volume of the base as follows:
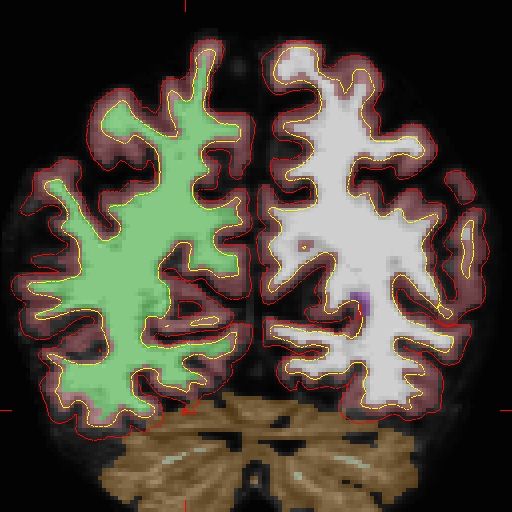
Below is a visualization of the problem area in the base: brain.finalsurfs.mgz, slice 89 at coordinates 109 166 89 in right hemi. The problem area extends through slices 85 - 91.
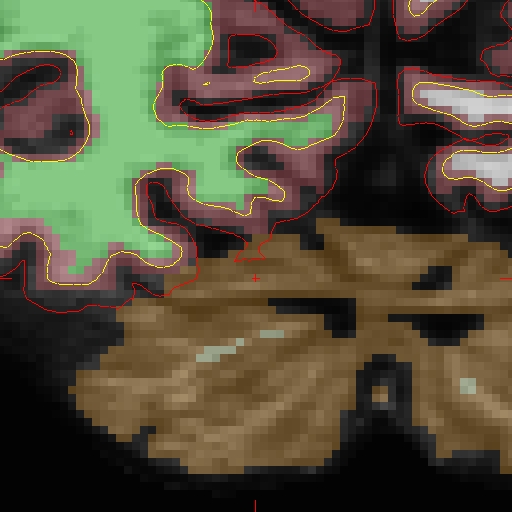
And here is the area zoomed in:
First make a copy of the brain.finalsurfs.mgz volume to one called 'brain.finalsurfs.manedit.mgz' and save it within the mri directory of the base:
cp OAS2_0002/mri/brain.finalsurfs.mgz OAS2_0002/mri/brain.finalsurfs.manedit.mgz
Next, load the brain.finalsurfs.manedit.mgz volume as the aux volume in tkmedit and edit this volume by going through the affected slices and removing the voxels in the cerebellum and surrounding areas that cause the pial surface misplacement. Once you are finished editing, save the changes to the volume with File – Save Aux volume. Then, run the following to have edits take effect:
recon-all -base OAS2_0002 -autorecon-pial
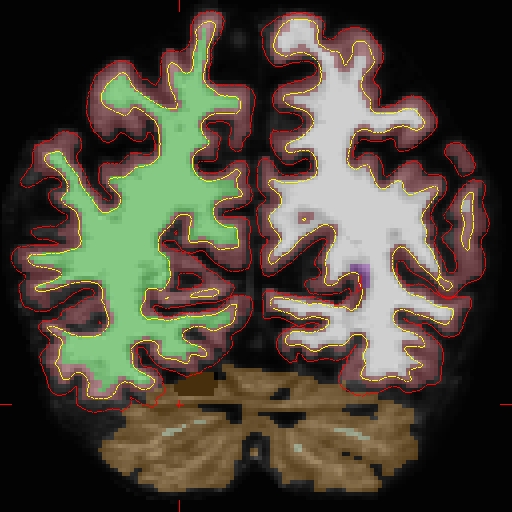
Results will look as follows (see OAS2_0002_fixed):
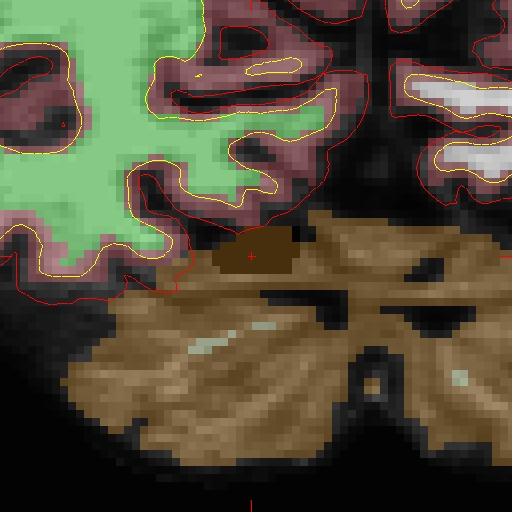
Here is zoomed in version of above:
Now that the base and cross sectional time point 2 are fixed, you must re-create the longitudinal data set for time point 2. Return to the Longitudinal Tutorial to read that section.
