|
Size: 5418
Comment:
|
← Revision 55 as of 2025-03-03 12:14:01 ⇥
Size: 10203
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| '''''This functionality is present in FreeSurfer 6.0 and later''''' | '''''We are working on a single Python program that unifies all the subregion segmentation modules; please see: [[https://surfer.nmr.mgh.harvard.edu/fswiki/SubregionSegmentation]].''''' '''''NEW: we have the atlas at the core of this module in MNI-ICBM space; please see [[https://surfer.nmr.mgh.harvard.edu/fswiki/SubfieldAtlasesICBMspace]].''''' |
| Line 7: | Line 9: |
| ''E-mail: iglesias [at] nmr.mgh.harvard.edu'' | ''E-mail: e.iglesias [at] ucl.ac.uk'' |
| Line 13: | Line 15: |
| * [[http://www.nmr.mgh.harvard.edu/~iglesias/pdf/Neuroimage_2015_brainstem.pdf|Bayesian segmentation of brainstem structures in MRI]]. Iglesias, J.E., Van Leemput, K., Bhatt, P., Casillas, C., Dutt, S., Schuff, N., Truran-Sacrey, D., Boxer, A., and Fischl, B. Neuroimage, in press. | * [[http://www.nmr.mgh.harvard.edu/~iglesias/pdf/Neuroimage_2015_brainstem.pdf|Bayesian segmentation of brainstem structures in MRI]]. Iglesias, J.E., Van Leemput, K., Bhatt, P., Casillas, C., Dutt, S., Schuff, N., Truran-Sacrey, D., Boxer, A., and Fischl, B. NeuroImage, 113, June 2015, 184-195. |
| Line 15: | Line 17: |
| See also: [[HippocampalSubfields]], [[LongitudinalHippocampalSubfields]], [[HippocampalSubfieldsAndNucleiOfAmygdala]], [[AANSegment]] | |
| Line 22: | Line 24: |
| 4. Frequently asked questions | 4. Gathering the volumes from all analyzed subjects 5. Frequently asked questions 6. Multimodal integration 7. Test data |
| Line 25: | Line 30: |
| <<BR>> | |
| Line 30: | Line 36: |
| Here is a sample segmentation produced by the algorithm. | Here are sample segmentations produced by the algorithm. |
| Line 37: | Line 43: |
| Line 40: | Line 45: |
| The brainstem module requires the Matlab R2012 runtime; the runtime is free, and therefore '''NO MATLAB LICENSES ARE REQUIRED TO USE THIS SOFTWARE.''' Please note that, if you have already installed the runtime for the hippocampal subfield module, you do not need to install it again. |
The brainstem module requires the Matlab R2012b (!FreeSurfer 6) or Matlab R2014b runtime (!FreeSurfer 7); these runtimes are free, and therefore '''NO MATLAB LICENSES ARE REQUIRED TO USE THIS SOFTWARE.''' Please note that, if you have already installed the runtime for the [[https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfieldsAndNucleiOfAmygdala|hippocampal subfield / amygdala module]] or the [[https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfields|FS6.0 subfield module]], you do not need to install it again. |
| Line 47: | Line 52: |
| <<BR>> | |
| Line 50: | Line 56: |
| To use this software, you simply need to add the flag -brainstem-structures to your recon-all command. For example, to analyze subject "bert", you would type: | In !FreeSurfer 6.0, you need to add the flag -brainstem-structures to your recon-all command. For example, to analyze subject "bert", you would type: |
| Line 62: | Line 68: |
| The output will be in the following directory: | In !FreeSurfer 7, this module is called independently from recon-all: |
| Line 64: | Line 70: |
| $SUBJECTS_DIR/bert/mri/brainstemAnalysis_v1.0 | {{{ segmentBS.sh bert [SUBJECTS_DIR] }}} where the second argument is optional, and can be used to override the !FreeSurfer subject directory. Bear in mind that the subject at hand ("bert" in this case) needs to have been processed with recon-all first (e.g., recon-all -all -s bert). |
| Line 67: | Line 77: |
| In this directory, you will find two files: | Both in !FreeSurfer 6 and 7, the output will be in the subjects mri directory, in this case: |
| Line 69: | Line 79: |
| ''discreteLabels.mgz'': the volume with the segmentation . | $SUBJECTS_DIR/bert/mri |
| Line 71: | Line 81: |
| ''volumesHippo.txt'': a text file with the volumes of the brainstem structures and of the whole brainstem, in cubic mm. | In this directory, you will find the following three files (where "v1x" is "v10" in !FreeSurfer 6, or v2 in !FreeSurfer 7): ''brainstemSsLabels.v1x.mgz'': the volume with the segmentation at 0.5 mm resolution. ''brainstemSsLabels.v1x.FSvoxelSpace.mgz'': discrete segmentation in the voxel space of the other FreeSurfer volumes (nu.mgz, aseg.mgz, etc). ''brainstemSsVolumes.v1x.txt'': a text file with the volumes of the brainstem structures and of the whole brainstem, in cubic mm. It will also create a file called brainstem.v1x.stats in the stats folder. This data can be compiled across subjects with the asegstats2table command, eg, {{{ asegstats2table --statsfile=brainstem.v12.stats --tablefile=brainstem.v12.dat ... }}} <<BR>> === 4. Gathering the volumes from all analyzed subjects === Once this module has been run on a number of subjects, it is possible to collect the volumes of the brainstem from all the subjects and write them to a single file - which can be easily read with a spreadsheet application. This can be done with ConcatenateSubregionsResults (please note that our older tool quantifyBrainstemStructures.sh is deprecated). |
| Line 74: | Line 101: |
| <<BR>> | |
| Line 75: | Line 103: |
| === 4. Frequently asked questions (FAQ) === | === 5. Frequently asked questions (FAQ) === |
| Line 81: | Line 110: |
| * '''The sum of the number of voxels of a given structure multiplied by the volume of a voxel is not equal to the volume reported in volumesBrainstem.txt.''' | * '''The sum of the number of voxels of a given structure multiplied by the volume of a voxel is not equal to the volume reported in brainstemSsVolumes.v1x.txt.''' |
| Line 83: | Line 112: |
| This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation in discreteLabels.mgz. This is the same that happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats. | This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation in brainstemSsLabels.v1x.mgz. This is the same that happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats. |
| Line 85: | Line 114: |
| * '''The size of the image volume of discreteLabels.mgz (in voxels) is not the same as that of norm.mgz or the additional input scan.''' | * '''The size of the image volume of brainstemSsLabels.v1x.mgz (in voxels) is not the same as that of norm.mgz or the additional input scan.''' |
| Line 87: | Line 116: |
| The segmentation discreteLabels.mgz covers only a patch around the brainstem, at a higher resolution than the input image. The segmentation and the image are defined in the same physical coordinates, so you can visualize them simultaneously with: | The segmentation brainstemSsLabels.v1x.mgz covers only a patch around the brainstem, at a higher resolution than the input image. The segmentation and the image are defined in the same physical coordinates, so you can visualize them simultaneously with: |
| Line 91: | Line 120: |
| freeview -v nu.mgz -v brainstemAnalysis_v1.0/discreteLabels.mgz:colormap=lut | freeview -v nu.mgz -v brainstemSsLabels.v1x.mgz:colormap=lut |
| Line 94: | Line 123: |
| If you want to resample the segmentation to the 1 mm FreeSurfer voxel space, you can run the command: | The software also produces brainstemSsLabels.v1x.FSvoxelSpace.mgz, in case you need a segmentation in the FreeSurfer voxel space (e.g., to create masks for subsequent analyses). * '''I have longitudinal data for a number of subjects. Is there a way of segmenting their brainstem structures in a longitudinal fashion?''' At this point, we have not tested or implemented any specific longitudinal segmentation methods for the brainstem structures. That said, if you process the subjects with the [[https://surfer.nmr.mgh.harvard.edu/fswiki/LongitudinalProcessing|main FreeSurfer longitudinal stream]], you can run the brainstem module on the longitudinally processed time points (rather than the cross-sectionally processed counterparts), which improves the robustness and sensitivity of the analysis, see for instance [[http://www.nmr.mgh.harvard.edu/~iglesias/pdf/Neuroimage_2016_longitudinal.pdf|this article]]. * '''I am interested in the soft segmentations (i.e., posterior probabilities), can I have access to them?''' Yes. All you need to do is to define an environment variable WRITE_POSTERIORS and set it to 1. For example, in tcsh or csh: |
| Line 96: | Line 133: |
| mri_convert brainstemAnalysis_v1.0/discreteLabels.mgz brainstemAnalysis_v1.0/discreteLabelsResampledT1.mgz -rt nearest -rl nu.mgz -odt float | setenv WRITE_POSTERIORS 1 |
| Line 98: | Line 135: |
| Or, in bash: {{{ export WRITE_POSTERIORS=1 }}} Then, the software will write a bunch of files under the subject's mri directory, with the format: ''posterior_<substructure>_v1x.mgz'' |
|
| Line 101: | Line 144: |
| Indeed! The deformation of the atlas towards the input scan(s) is parallelized. If you want to limit the number of cores that the software is allowed to use, you simply need to set the environment variable ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS. For example, to limit the number of used cores to 2 in tcsh or csh: | Indeed! The deformation of the atlas towards the input scan(s) is parallelized, however, it will only use one core by default. If you want to use more than one core / thread, you can do that through the environment variable ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS. For example, to use two cores (tcsh/csh): |
| Line 105: | Line 149: |
| Or, in bash: | Or, in bash: |
| Line 113: | Line 157: |
* (Only applies to FreeSurfer 6.0) '''I ran recon-all -all -brainstem-structures and, after running for hours, the code stopped because I had not installed the Matlab runtime. Do I need to rerun everything?''' No! You can just run recon-all -brainstem-structures (that is, without the -all). Of course, that would be after installing the runtime. <<BR>> === 6. Multimodal integration === To extract values within the subfields from another modality (eg, DTI or fMRI), use vol2subfield. Run with --help for examples. vol2subfield is not in FS version 6, but you can get it from https://surfer.nmr.mgh.harvard.edu/pub/dist/freesurfer/6.0.0-patch/vol2subfield === 7. Test data === Processed data of a single subject (also used to demo the hippocampal subfield segmentation) is available here: {{{ wget -c ftp://surfer.nmr.mgh.harvard.edu/pub/data/hipposubfields-testdata.tgz }}} The data is extracted with: {{{ tar zxvf hipposubfields-testdata.tgz }}} Note: within the Martinos Center, this data is found here, but should not be modified (make a copy) as it is used for testing: {{{ /autofs/cluster/freesurfer/subjects/test/hipposubfields }}} The data is one subject from the ADNI dataset, is fully recon-d. The commands used to produce the brainstem data was: {{{ recon-all -s ADNI_135_S_4863_20-Feb-2013 segmentBS.sh ADNI_135_S_4863_20-Feb-2013 }}} To view the data: {{{ cd hipposubfields/ADNI_135_S_4863_20-Feb-2013/mri freeview -v nu.mgz -v brainstemSsLabels.v12.mgz:colormap=lut }}} |
Brainstem Substructures
We are working on a single Python program that unifies all the subregion segmentation modules; please see: https://surfer.nmr.mgh.harvard.edu/fswiki/SubregionSegmentation.
NEW: we have the atlas at the core of this module in MNI-ICBM space; please see https://surfer.nmr.mgh.harvard.edu/fswiki/SubfieldAtlasesICBMspace.
Author: Juan Eugenio Iglesias
E-mail: e.iglesias [at] ucl.ac.uk
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use these tools in your analysis, please cite:
Bayesian segmentation of brainstem structures in MRI. Iglesias, J.E., Van Leemput, K., Bhatt, P., Casillas, C., Dutt, S., Schuff, N., Truran-Sacrey, D., Boxer, A., and Fischl, B. NeuroImage, 113, June 2015, 184-195.
See also: HippocampalSubfields, LongitudinalHippocampalSubfields, HippocampalSubfieldsAndNucleiOfAmygdala, AANSegment
Contents
- Motivation and General Description
- Installation
- Usage
- Gathering the volumes from all analyzed subjects
- Frequently asked questions
- Multimodal integration
- Test data
1. Motivation and General Description
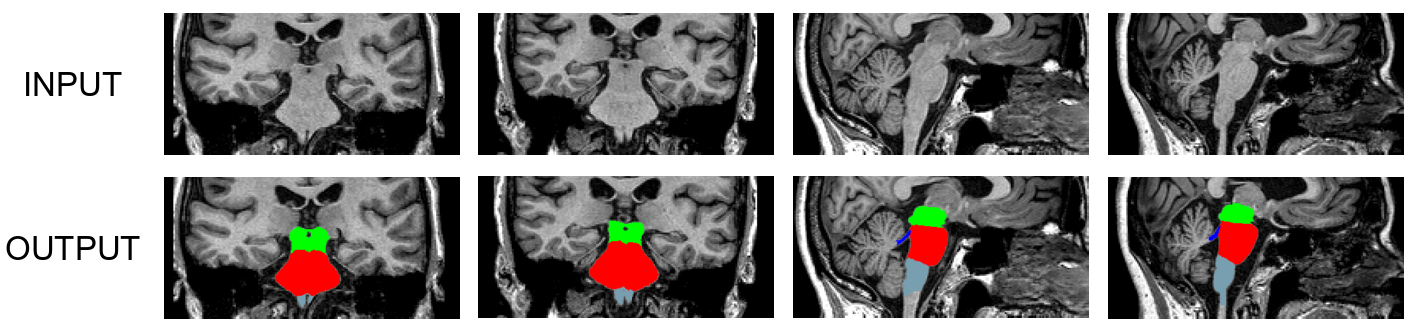
This tool generates an automated segmentation of four different brainstem structures from the input T1 scan: medulla oblongata, pons, midbrain and superior cerebellar peduncle (SCP). We use a Bayesian segmentation algorithm that relies on a probabilistic atlas of the brainstem (and neighboring brain structures) built upon manual delineations of the structures on interest in 49 scans (10 for the brainstem structures, 39 for the surrounding structures). The delineation protocol for the brainstem was designed by Dr. Adam Boxer and his team at the UCSF Memory and Aging Center, and is described in the paper.
Here are sample segmentations produced by the algorithm.
2. Installation
The brainstem module requires the Matlab R2012b (FreeSurfer 6) or Matlab R2014b runtime (FreeSurfer 7); these runtimes are free, and therefore NO MATLAB LICENSES ARE REQUIRED TO USE THIS SOFTWARE. Please note that, if you have already installed the runtime for the hippocampal subfield / amygdala module or the FS6.0 subfield module, you do not need to install it again. Instructions for the installation of the runtime can be found here:
https://surfer.nmr.mgh.harvard.edu/fswiki/MatlabRuntime
3. Usage
In FreeSurfer 6.0, you need to add the flag -brainstem-structures to your recon-all command. For example, to analyze subject "bert", you would type:
recon-all -all -s bert -brainstem-structures
Or, if Bert has already undergone the FreeSurfer pipeline (recon-all -all), you can just run:
recon-all -s bert -brainstem-structures
In FreeSurfer 7, this module is called independently from recon-all:
segmentBS.sh bert [SUBJECTS_DIR]
where the second argument is optional, and can be used to override the FreeSurfer subject directory. Bear in mind that the subject at hand ("bert" in this case) needs to have been processed with recon-all first (e.g., recon-all -all -s bert).
Both in FreeSurfer 6 and 7, the output will be in the subjects mri directory, in this case:
$SUBJECTS_DIR/bert/mri
In this directory, you will find the following three files (where "v1x" is "v10" in FreeSurfer 6, or v2 in FreeSurfer 7):
brainstemSsLabels.v1x.mgz: the volume with the segmentation at 0.5 mm resolution.
brainstemSsLabels.v1x.FSvoxelSpace.mgz: discrete segmentation in the voxel space of the other FreeSurfer volumes (nu.mgz, aseg.mgz, etc).
brainstemSsVolumes.v1x.txt: a text file with the volumes of the brainstem structures and of the whole brainstem, in cubic mm.
It will also create a file called brainstem.v1x.stats in the stats folder. This data can be compiled across subjects with the asegstats2table command, eg,
asegstats2table --statsfile=brainstem.v12.stats --tablefile=brainstem.v12.dat ...
4. Gathering the volumes from all analyzed subjects
Once this module has been run on a number of subjects, it is possible to collect the volumes of the brainstem from all the subjects and write them to a single file - which can be easily read with a spreadsheet application. This can be done with ConcatenateSubregionsResults (please note that our older tool quantifyBrainstemStructures.sh is deprecated).
5. Frequently asked questions (FAQ)
Are you sure that this software does not require Matlab licenses? Why does it require the Matlab runtime, then?
The software uses compiled Matlab code that requires the free runtime, but no licenses. So, if you have a computer cluster, you can run hundred of segmentations in parallel without having to worry about Matlab licenses. And yes, this is all perfectly legal ![]()
The sum of the number of voxels of a given structure multiplied by the volume of a voxel is not equal to the volume reported in brainstemSsVolumes.v1x.txt.
This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation in brainstemSsLabels.v1x.mgz. This is the same that happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats.
The size of the image volume of brainstemSsLabels.v1x.mgz (in voxels) is not the same as that of norm.mgz or the additional input scan.
The segmentation brainstemSsLabels.v1x.mgz covers only a patch around the brainstem, at a higher resolution than the input image. The segmentation and the image are defined in the same physical coordinates, so you can visualize them simultaneously with:
cd $SUBJECTS_DIR/<subject_name>/mri/ freeview -v nu.mgz -v brainstemSsLabels.v1x.mgz:colormap=lut
The software also produces brainstemSsLabels.v1x.FSvoxelSpace.mgz, in case you need a segmentation in the FreeSurfer voxel space (e.g., to create masks for subsequent analyses).
I have longitudinal data for a number of subjects. Is there a way of segmenting their brainstem structures in a longitudinal fashion?
At this point, we have not tested or implemented any specific longitudinal segmentation methods for the brainstem structures. That said, if you process the subjects with the main FreeSurfer longitudinal stream, you can run the brainstem module on the longitudinally processed time points (rather than the cross-sectionally processed counterparts), which improves the robustness and sensitivity of the analysis, see for instance this article.
I am interested in the soft segmentations (i.e., posterior probabilities), can I have access to them?
Yes. All you need to do is to define an environment variable WRITE_POSTERIORS and set it to 1. For example, in tcsh or csh:
setenv WRITE_POSTERIORS 1
Or, in bash:
export WRITE_POSTERIORS=1
Then, the software will write a bunch of files under the subject's mri directory, with the format: posterior_<substructure>_v1x.mgz
This module is CPU hungry
Indeed! The deformation of the atlas towards the input scan(s) is parallelized, however, it will only use one core by default. If you want to use more than one core / thread, you can do that through the environment variable ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS. For example, to use two cores (tcsh/csh):
setenv ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS 2
Or, in bash:
export ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS=2
The volume of the whole brainstem obtained with this module is not equal to the value reported by the main FreeSurfer pipeline in $SUBJECTS_DIR/<subject_name>/stats/aseg.stats.
Yes! There are two reasons for this. First, the volumes correspond to two different analyses. And second: the anatomical definitions of the brainstem are (particularly in the midbrain) are different in the original FreeSurfer protocol and the UCSF protocol used by this module.
(Only applies to FreeSurfer 6.0) I ran recon-all -all -brainstem-structures and, after running for hours, the code stopped because I had not installed the Matlab runtime. Do I need to rerun everything?
No! You can just run recon-all -brainstem-structures (that is, without the -all). Of course, that would be after installing the runtime.
6. Multimodal integration
To extract values within the subfields from another modality (eg, DTI or fMRI), use vol2subfield. Run with --help for examples. vol2subfield is not in FS version 6, but you can get it from https://surfer.nmr.mgh.harvard.edu/pub/dist/freesurfer/6.0.0-patch/vol2subfield
7. Test data
Processed data of a single subject (also used to demo the hippocampal subfield segmentation) is available here:
wget -c ftp://surfer.nmr.mgh.harvard.edu/pub/data/hipposubfields-testdata.tgz
The data is extracted with:
tar zxvf hipposubfields-testdata.tgz
Note: within the Martinos Center, this data is found here, but should not be modified (make a copy) as it is used for testing:
/autofs/cluster/freesurfer/subjects/test/hipposubfields
The data is one subject from the ADNI dataset, is fully recon-d. The commands used to produce the brainstem data was:
recon-all -s ADNI_135_S_4863_20-Feb-2013 segmentBS.sh ADNI_135_S_4863_20-Feb-2013
To view the data:
cd hipposubfields/ADNI_135_S_4863_20-Feb-2013/mri freeview -v nu.mgz -v brainstemSsLabels.v12.mgz:colormap=lut
