|
Size: 2284
Comment:
|
Size: 2008
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [[fswiki|top]] | |
| Line 4: | Line 3: |
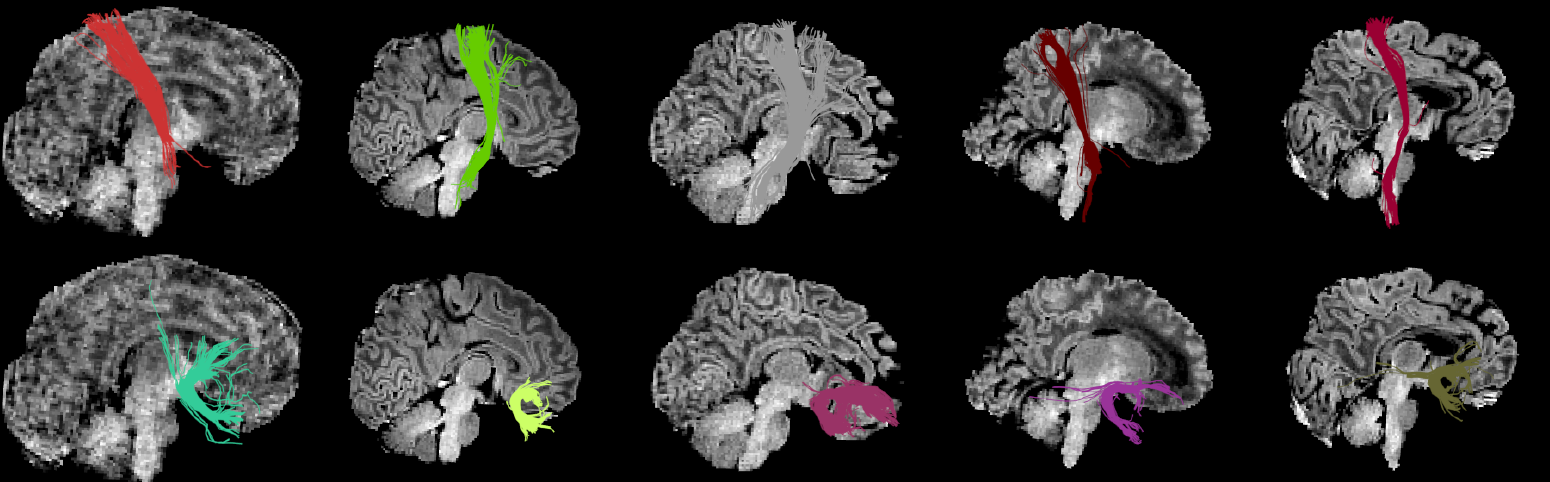
| This method finds corresponding clusters across subjects. Currently only one-to-one cluster's correspondences are available using the Hungarian algorithm. | This method finds corresponding clusters across subjects. Currently, only one-to-one cluster's correspondences are available using the Hungarian algorithm. Importantly, by using our anatomical similarity metric, correspondences are found without the need of registration comparing clusters of streamlines from each subject's native space. {{attachment:hungarian_babies.png||height="360px"}}{ |
| Line 9: | Line 10: |
| A use case analysis merely addresses the most obvious of questions: who needs the software, and what are they going to do with it. | The Hungarian algorithm finds corresponding clusters between two subjects. |
| Line 11: | Line 12: |
| === Actors (Users) === In UML terminology, the persons (or software agents) external to a software component are called the actors. |
{{{ AnatomiCuts_correspondences -s1 segmentation1.nii.gz -s2 segmentation2.nii.gz -c numClusters -h1 clusteringPath1 -h2 clusteringPath2 -m metric -o output.csv }}} |
| Line 14: | Line 16: |
| ==== Research ==== === Scenarios (Use Cases) === These establish the framework for test cases. They also bring out the vocabulary of the system. This vocabulary is defined in the Terms section following these scenarios. |
Where |
| Line 18: | Line 18: |
| ==== Use Case #1 ==== ==== Use Case #2 ==== ==== Use Case #3 ==== ==== Use Case #4 ==== === Terms === The following is a list of some of the vocabulary used in the preceding scenarios, plus terms that are common across the system in which the software is used. |
-s1 the segmentation to be used for anatomical similarity in subject one. |
| Line 25: | Line 20: |
| == Requirements == === General Requirements === === Specific Requirements === == Implementation == === API === === System Architecture and Primary Components === === Classes === === Collaboration and Sequence Diagrams === UML diagrams describing the time-course of the objects composing the executable. |
-s2 the segmentation to be used for anatomical similarity in subject one. |
| Line 35: | Line 22: |
| === Properties === Here are listed the configurable properties of the executable. These can be configurables read from a configuration file, or configurables hard-coded into the source code. |
-h1 the path to the AnatomiCuts folder to be used for subject1. |
| Line 38: | Line 24: |
| == Test Plan == === Introduction === Tests should cover the following categories of testing. |
-h2 the path to the AnatomiCuts folder to be used for subject2. |
| Line 42: | Line 26: |
| ==== Functional ==== This type of test ascertains whether the software executes its basic functionality under optimal conditions. |
-m metric to be used: labels (anatomical similarity) or euclid (euclidean similarity). |
| Line 45: | Line 28: |
| ==== Boundary ==== This type of test determines the breaking points of the software, and whether the software gracefully handles input near and beyond these boundaries. |
-sym (under development) this flag will mirror the segmentation in subject 2 to find between hemisphere correspondences. |
| Line 48: | Line 30: |
| ==== Stability ==== This type of test determines long-term behavior of the software: whether is has a memory leak, or prone to crashes which are not repeatable in any single run of any of the other tests. |
-o output csv file |
| Line 51: | Line 32: |
| ==== Performance ==== These tests produce benchmarks on the performance of the software. |
== Output == The output will be a csv file: {{{ Subject A, Subject B 100000,11111 1010101, 100010 }}} Where cluster 100000.trk from Subject A corresponds to cluster 11111.trk from Subject B |
| Line 55: | Line 49: |
V. Siless, J. Y. Davidow, J. Nielsen, Q. Fan, T. Hedden, M. Hollinshead, C. V. Bustamante, M. K. Drews, K. R. A. Van Dijk, M.A. Sheridan, R. L. Buckner, B. Fischl, L. Somerville, and A. Yendiki. 2017. “Registration-free analysis of diffusion MRI tractography data across subjects through the human lifespan.” V. Siless, K. Chang, B. Fischl, and A. Yendiki. 2018. “AnatomiCuts: Hierarchical clustering of tractography streamlines based on anatomical similarity.” NeuroImage, 166, Pp. 32-45. |
AnatomiCuts correspondences
This method finds corresponding clusters across subjects. Currently, only one-to-one cluster's correspondences are available using the Hungarian algorithm. Importantly, by using our anatomical similarity metric, correspondences are found without the need of registration comparing clusters of streamlines from each subject's native space.
 {
{
The Hungarian algorithm
The Hungarian algorithm finds corresponding clusters between two subjects.
AnatomiCuts_correspondences -s1 segmentation1.nii.gz -s2 segmentation2.nii.gz -c numClusters -h1 clusteringPath1 -h2 clusteringPath2 -m metric -o output.csv
Where
-s1 the segmentation to be used for anatomical similarity in subject one.
-s2 the segmentation to be used for anatomical similarity in subject one.
-h1 the path to the AnatomiCuts folder to be used for subject1.
-h2 the path to the AnatomiCuts folder to be used for subject2.
-m metric to be used: labels (anatomical similarity) or euclid (euclidean similarity).
-sym (under development) this flag will mirror the segmentation in subject 2 to find between hemisphere correspondences.
-o output csv file
Output
The output will be a csv file:
Subject A, Subject B 100000,11111 1010101, 100010
Where cluster 100000.trk from Subject A corresponds to cluster 11111.trk from Subject B
References
V. Siless, J. Y. Davidow, J. Nielsen, Q. Fan, T. Hedden, M. Hollinshead, C. V. Bustamante, M. K. Drews, K. R. A. Van Dijk, M.A. Sheridan, R. L. Buckner, B. Fischl, L. Somerville, and A. Yendiki. 2017. “Registration-free analysis of diffusion MRI tractography data across subjects through the human lifespan.”
V. Siless, K. Chang, B. Fischl, and A. Yendiki. 2018. “AnatomiCuts: Hierarchical clustering of tractography streamlines based on anatomical similarity.” NeuroImage, 166, Pp. 32-45.
